Numpy is one of the core modules of the Scipy “ecosystem”;
however, additional numerical
routines are packages in the separate scipy module.
The sub-modules contained in scipy include:
scipy.cluster : Clustering and cluster analysisscipy.fftpack : Fast fourier transform functionsscipy.integrate : Integration and ODE (Ordinary Differential Equations)scipy.interpolate : Numerical interpolation including splines and radial basis function interpolationscipy.io : Input/Output functions for e.g. Matlab (.mat) files, wav (sound) files, and netcdf.scipy.linalg : Linear algebra functions like eigenvalue analysis, decompositoins, and matrix equation solversscipy.ndimage : Image processing functions (works on high dimensional images - hence nd!)scipy.optimize : Optimization (minimize, maximize a function wrt variables), curve fitting, and root findingscipy.signal : Signal processing (1d) including convolution, filtering and filter design, and spectral analysisscipy.sparse : Sparse matrix support (2d)scipy.spatial : Spatial analysis related functionsscipy.statistical : Statistical analysis including continuous and discreet distributions and a slew of statistical functionsWhile we don’t have time to cover all of these submodules in detail, we will give a introduction to their general usage with a couple of exercise and then a sample project using a few of them to perform some 1d signal processing.
Curve fitting represents a relatively common research task, and fortunatley Python includes some great curve fitting functionalities.
Some of the more advanced ones are present in additional Scikit libraries,
but Scipy includes some of the most common algorithms in the optimize
submodule, as well as the odr submodule, which covers the more elaborate
“orthogonal distance regression” (as opposed to ordinary least squares
used in curve_fit and leastsq).
Load the data contained in the following file
and use the scipy.optimize.curve_fit or scipy.optimize.leastsq (least-squares)
fitting function (curve_fit calls leastsq but doesn’t give as much output!)
to generate a line of best fit for the data.
You shold evaluate several possible functions for the fit including
f(x,a,b) = a*x + bf(x,a,b,c) = a*x^2 + b*x + cf(x,a,b,c) = a*e^(b*x) + cGenerate an estimate of the goodness of fit to discriminate between these possible models.
Bonus section
Try the same again with the following data (you should be able to just change the filename!)
You should find that the goodness of fit has become worse. In fact the algorithm used
by Scipy’s optimize submodule is not always able to determine which of the models fits best.
Try using the scipy.odr submodule instead; a guide for how to do this can be found
here
General tips:
lambdas used here,
or stick with defining the functions the usual way, with def).import numpy as np
import scipy.optimize as scopt
def perform_fitting(filename):
funcs = {
'linear' : lambda x,a,b,c : a*x + b,
'quadratic' : lambda x,a,b,c : a*x*x + b*x + c,
'exponential' : lambda x,a,b,c : a*np.exp(b*x) + c,
}
data = np.fromfile(filename)
Npoints = len(data)
taxis = np.arange(Npoints)
for fname in funcs:
popt, pcov = scopt.curve_fit( funcs[fname], taxis, data)
yexact = funcs[fname](taxis, *popt)
residuals = data - yexact
ss_err = (residuals**2).sum()
ss_tot = ((data-data.mean())**2).sum()
rsquared = 1-(ss_err/ss_tot)
print("\n\n\nFitting: %s"%fname)
print("\nCurve fit")
print("popt:", popt)
print("pcov:", pcov)
print("rsquared:", rsquared)
if __name__ == "__main__":
perform_fitting("curve_fitting_data_simple.txt")
The first part of the quesrtion generates the output below (the last line represents the R^2 goodness of fit statistic for each fit)
Fitting: quadratic
Curve fit
popt: [ 0.27742012 0.43227489 24.42625192]
pcov: [[ 1.24062405e-04 -2.35718583e-03 7.07155749e-03]
[ -2.35718583e-03 4.80617819e-02 -1.65474460e-01]
[ 7.07155749e-03 -1.65474460e-01 8.07571966e-01]]
rsquared: 0.99839145357
Fitting: linear
Curve fit
popt: [ 5.70325725 8.61330482 1. ]
pcov: inf
rsquared: 0.939693857665
Fitting: exponential
Curve fit
popt: [ 19.98312703 0.10003282 2.07108578]
pcov: [[ 2.35952209e-03 -5.30270860e-06 -3.10538435e-03]
[ -5.30270860e-06 1.20524441e-08 6.87521073e-06]
[ -3.10538435e-03 6.87521073e-06 4.24287754e-03]]
rsquared: 0.999998872202
We see from this that for non-noisy data using curve_fit performs relatively
well (i.e. the data was indeed exponential!).
If we run the same function on the file “curve_fitting_data_noisy.txt”, we get
Fitting: quadratic
Curve fit
popt: [ 0.36402064 1.39338512 33.74952291]
pcov: [[ 1.16491630e-02 -2.21334100e-01 6.64002296e-01]
[ -2.21334100e-01 4.51288594e+00 -1.55376545e+01]
[ 6.64002296e-01 -1.55376545e+01 7.58290698e+01]]
rsquared: 0.932781903212
Fitting: linear
Curve fit
popt: [ 8.30977729 13.0003464 1. ]
pcov: inf
rsquared: 0.887804486031
Fitting: exponential
Curve fit
popt: [ 37.66928574 0.08831645 -6.97576507]
pcov: [[ 5.63753689e+02 -6.41775284e-01 -6.99939382e+02]
[ -6.41775284e-01 7.38443618e-04 7.87263594e-01]
[ -6.99939382e+02 7.87263594e-01 8.90820497e+02]]
rsquared: 0.933020625779
i.e. as far as curve_fit is concerned, both quadratic and exponential are relatively good fits,
and the one that “wins out” will vary depending on the noise present.
However, if we add in the additional odrpack version of the stats
by adding
import scipy.odr as scodr
by the import statements, and the following to the loop over functions:
...
print("\nODRPACK VERSION")
funcnow = lambda params, x: funcs[fname](x, *params)
mod = scodr.Model(funcnow)
dat = scodr.Data(taxis,data)
odr = scodr.ODR(dat, mod, beta0 = [1.0, 1.0, 0.0])
out = odr.run()
out.pprint()
we also produce
Fitting: quadratic
...
ODRPACK VERSION
Beta: [ 0.55201606 -0.63943926 31.30145991]
Beta Std Error: [ 0.08980006 1.10407977 1.93744129]
Beta Covariance: [[ 0.00241881 -0.0263813 0.01923865]
[-0.0263813 0.36563691 -0.35584366]
[ 0.01923865 -0.35584366 1.12591663]]
Residual Variance: 3.333886943328627
Inverse Condition #: 0.0020568238964839263
Reason(s) for Halting:
Iteration limit reached
Fitting: linear
...
ODRPACK VERSION
Beta: [ 9.34657515 3.1507637 0. ]
Beta Std Error: [ 0.78204438 8.56164287 0. ]
Beta Covariance: [[ 0.14923625 -1.41774486 0. ]
[ -1.41774486 17.8865157 0. ]
[ 0. 0. 0. ]]
Residual Variance: 4.098155829657788
Inverse Condition #: 0.0437541938307872
Reason(s) for Halting:
Problem is not full rank at solution
Sum of squares convergence
Fitting: exponential
...
ODRPACK VERSION
Beta: [ 43.9540789 0.08427052 -17.18528396]
Beta Std Error: [ 2.68421883e+01 2.80794795e-02 3.04612945e+01]
Beta Covariance: [[ 2.69641542e+02 -2.79919119e-01 -3.04410441e+02]
[ -2.79919119e-01 2.95072729e-04 3.13666736e-01]
[ -3.04410441e+02 3.13666736e-01 3.47254336e+02]]
Residual Variance: 2.672077403678681
Inverse Condition #: 8.544672221858744e-06
Reason(s) for Halting:
Sum of squares convergence
i.e. we see that the residual variance and inverse condition are significantly lower for the exponential fitting (and these values are also more robust to noise).
Another common use of Scipy is signal interpolation.
Background : Interpolation
Interpolation is often used to resample data, generating data points at a finer spacing than the original data.
A simple example of interpolation is image resizing; when a small image is resized to a larger size, the “in-between” pixel values are generated by interpolating their values based on the nearest original pixel values.
Typical interpolation schemes include nearest, linear, and spline interpolation of various degrees. These terms denote the functional form of the interpolating function.
- nearest neighbor interpolation assigns each interpolated value the value of the nearest original value
- linear interpolation assigns each interpolated value the linearly weighted combination of the neighbouring values; e.g. if the value to be interpolated is 9 times closer to original point B than original point A, then the final interpolated value would be 0.1A + 0.9B.
- Spline interpolation approximates the region between the original data points as quadratic or cubic splines (depending on which spline interpolation order) was selected.
Download the data from here:
and use “linear” interpolation to interpolate the data to increase so that the new “x axis” has five times as many points between then original limits (i.e. you’ll resample the data to 5 times the density).
Run the interpolation with “quadratic” and “cubic” interpolation also (it helps if your interpolation code was in a function!), and you should see much better results!
Use the Numpy load function to load the data (as it was created with save!).
If you plot the data ( data[0] vs data[1] ), you should see a jagged sine curve.
Use the linspace function to create your new, denser x axis data.
After interpolation, you should end up with a slightly smoother sine curve.
import scipy.interpolate as scint
import numpy as np
import matplotlib.pyplot as plt # For plotting
def interp_data(data, kind="quadratic"):
plt.plot(data[0], data[1])
func = scint.interp1d(data[0], data[1], kind=kind)
xnew = np.linspace(data[0][0], data[0][-1], num=5*len(data[0]))
ynew = func(xnew)
plt.plot(xnew, ynew, '-r')
plt.title("Using %s interpolation"%kind)
plt.show()
if __name__ == '__main__':
data= np.load("data_interpolation.npy")
for kind in ["linear", "quadratic", "cubic"]:
interp_data(data, kind=kind)
will produce 3 plots to the screen; the cubic and quadratic version of the interpolation should look much smoother (closer to a real sine curve).
As you may not have a background in signal processing, as our last exercise, we’ll you through the process instead of leaving you to generate the code from scratch.
Download the wav file (uncompressed audio) from here Wav File.
This sound file will be the basis of our experimentation with some of the additional Scipy modules
scipy.ioscipy.io contains file reading functionality to load Matlab .mat files,
NetCDF, and wav audio files, amongst others.
We will use the wavfile submodule to load our audio file;
import scipy.io.wavfile as scwav
data, rate = scwav.read("audio_sample.wav")
Next we can inspect the the loaded data using a plot (plotting just the first 500 values),
import matplotlib.pyplot as plt
# Create a "time" axis array; at present there are
# `rate` data-points per second, as rate corresponds to the
# data sample rate.
Nsamples = len(data)
time_axis = np.arange(Nsamples)/rate
plt.plot( time_axis[:500], data[:500])
plt.show()
We should see an audio trace that looks something like
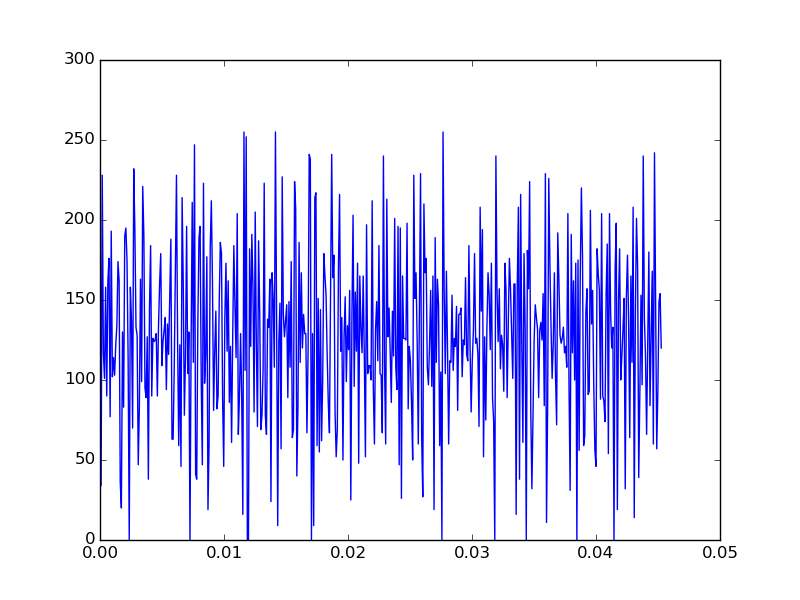
If you have headphones with you and play the audio, you’ll hear a noisy, but still recongnizable sound-bite.
Let’s see if we can improve the sound quality by performing some basic signal processing!
scipy.signalThere are a host of available 1d filters available in Scipy.
We’re going to construct a simple low-pass filter using the
firwin function;
import scipy.signal as scsig
# Calculate the Nqyist rate
nyq_rate = rate / 2.
# Set a low cutoff frequency of the filter: 1KHz
cutoff_hz = 1000.0
# Length of the filter (number of coefficients, i.e. the filter order + 1)
numtaps = 29
# Use firwin to create a lowpass FIR filter
fir_coeff = scsig.firwin(numtaps, cutoff_hz/nyq_rate)
# Use lfilter to filter the signal with the FIR filter
filtered = scsig.lfilter(fir_coeff, 1.0, data)
# Write the filtered version into an audio file so we can see
# if we can hear a difference!
scwav.write("filtered.wav", rate, filtered)
While “manual” verification by listening to the filtered signal can be useful, we would probably want to also analyze both the original signal and the effect the filtering has had.
scipy.fftpack and scipy.signalTo analyze a 1d filter, we often generate a periodogram, which essentially gives us information about the frequency content of the signal. The perriodogram itself is a power-spectrum representation of the Fourier transform of the signal; however, this is not a detailed course in 1d signal processing, so if you have no idea what that means then don’t worry about it!
For sound signals, the periodogram relates directly to the pitches of the sounds in the signal.
Let’s generate and plot a periodogram using scipy:
plt.figure()
f, Pxx = scsig.periodogram(data.astype(float), rate)
plt.semilogy(f, Pxx)
plt.xlabel('frequency [Hz]')
plt.ylabel('PSD [V**2/Hz]')
plt.show()
plt.figure()
f, Pxx = scsig.periodogram(filtered.astype(float), rate)
plt.semilogy(f, Pxx)
plt.xlabel('frequency [Hz]')
plt.ylabel('PSD [V**2/Hz]')
plt.show()
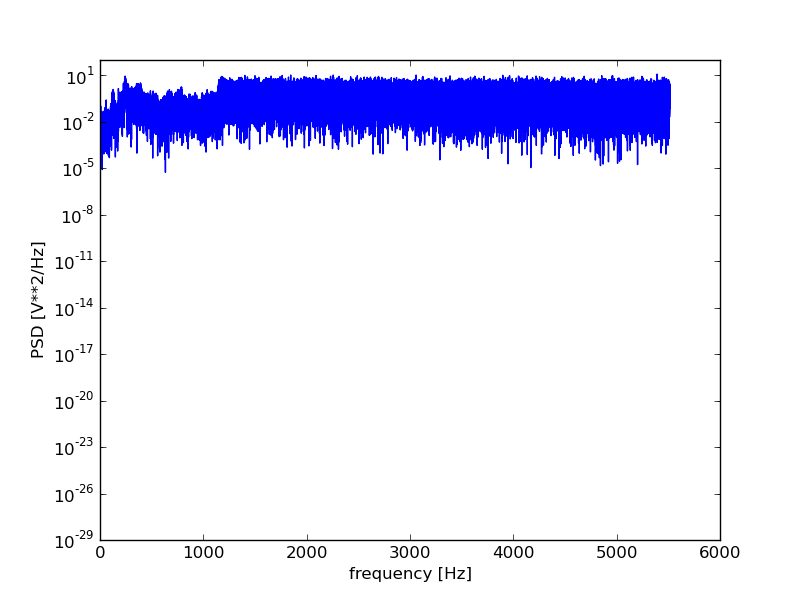
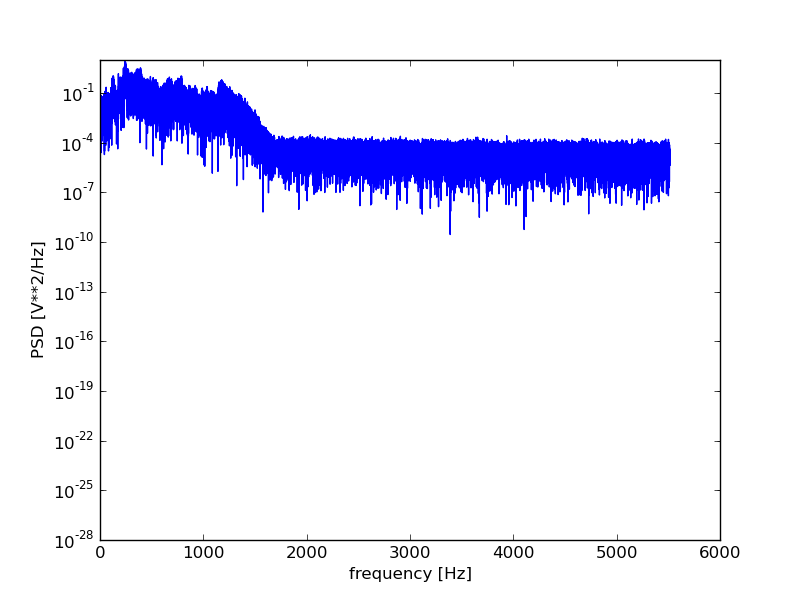
For those wanting more control over the spectral analysis, the fftpack
submodule offers access to a range of Fast-Fourier transform related
functions.
Performing the spectral analysis shows us that the signal did indeed contain a high frequency noise component that the filtering should, at least to some degree have helped remove.
If you would like to further practice Scipy and it’s submodules, please visit this resource.