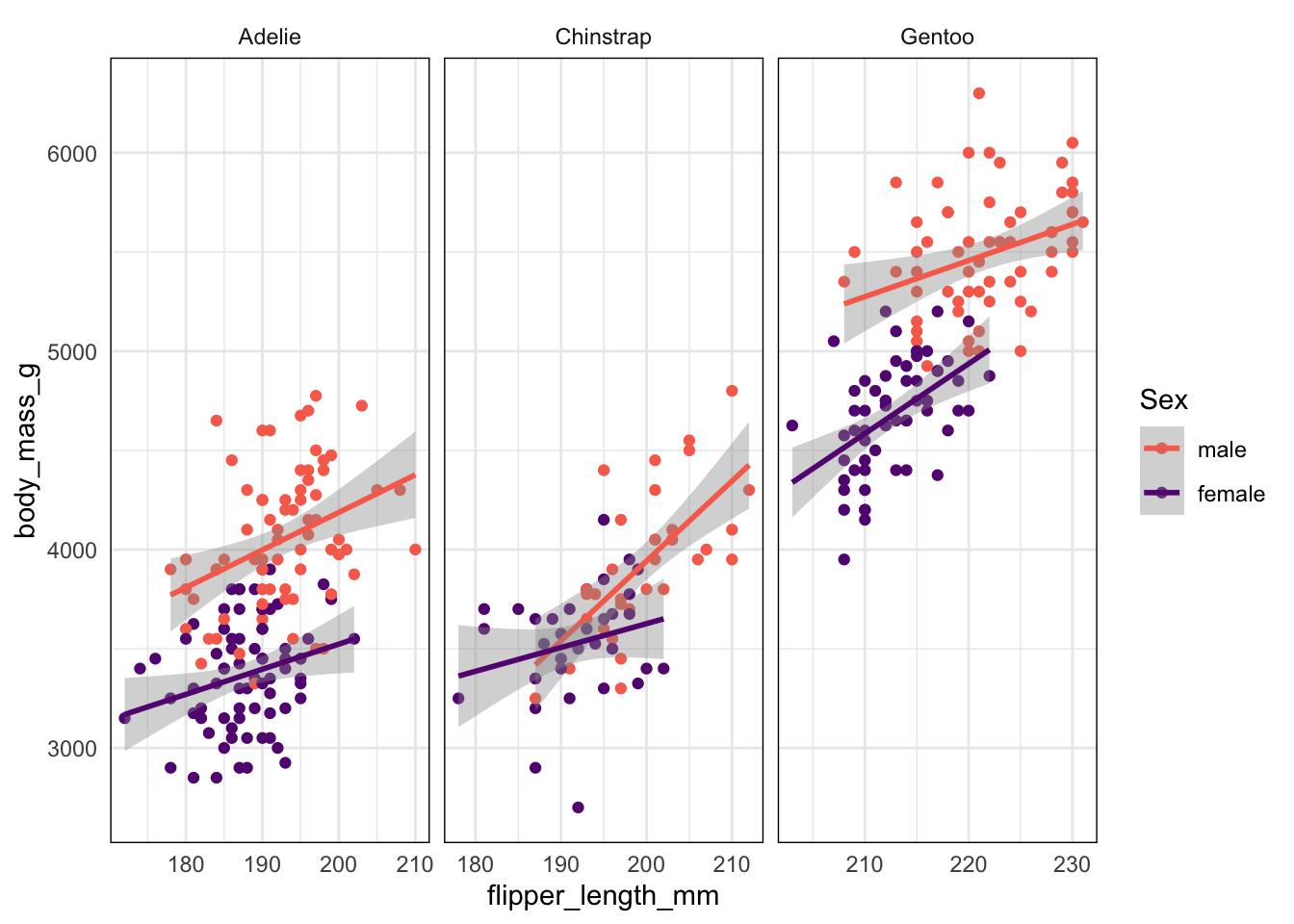
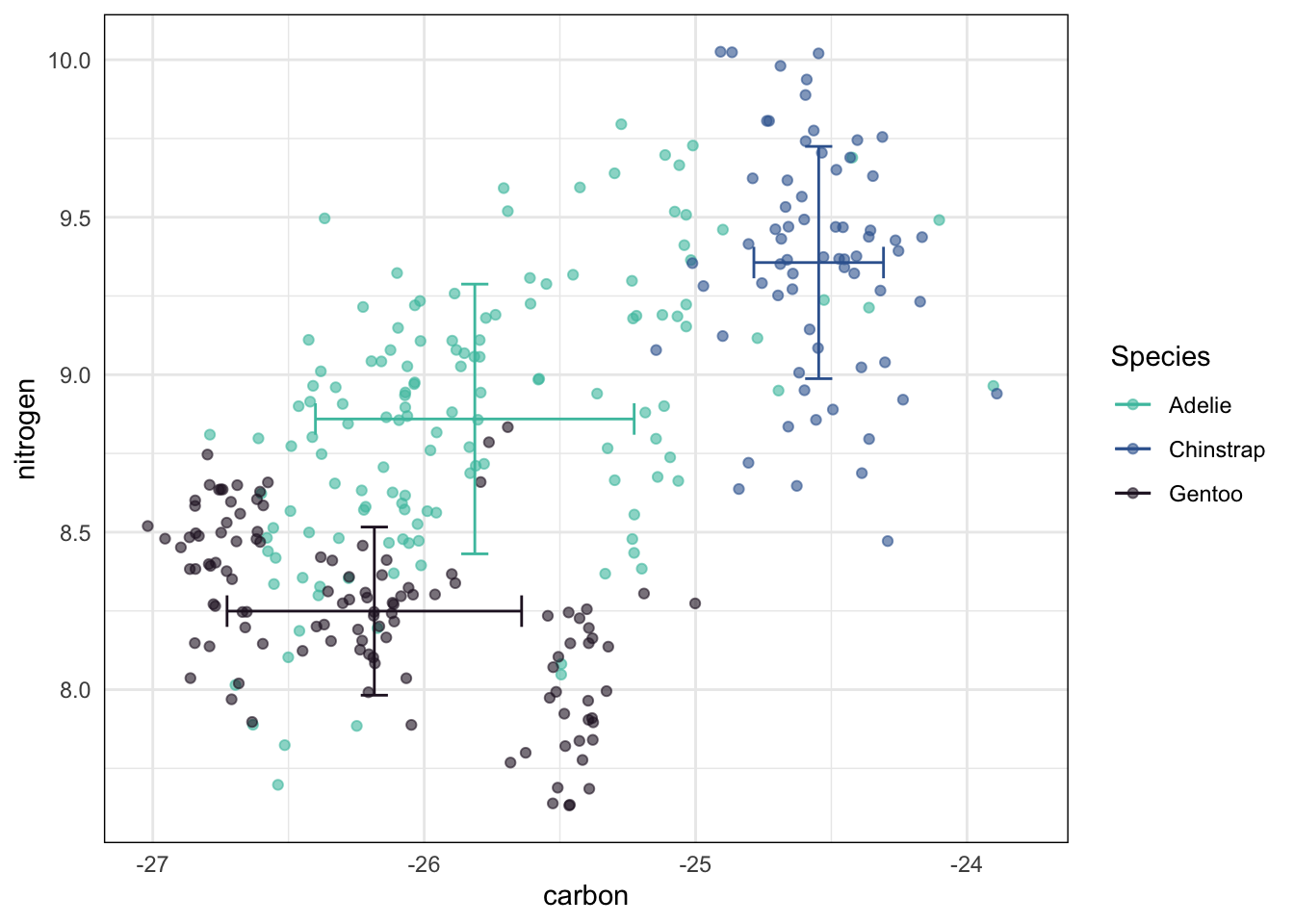
Rows: 333
Columns: 22
$ study_name <chr> "PAL0708", "PAL0708", "PAL0708", "PAL0708", "PAL0708…
$ sample_number <dbl> 1, 2, 3, 5, 6, 7, 8, 13, 14, 15, 16, 17, 18, 19, 20,…
$ species <chr> "Adelie", "Adelie", "Adelie", "Adelie", "Adelie", "A…
$ region <chr> "Anvers", "Anvers", "Anvers", "Anvers", "Anvers", "A…
$ island <chr> "Torgersen", "Torgersen", "Torgersen", "Torgersen", …
$ stage <chr> "Adult, 1 Egg Stage", "Adult, 1 Egg Stage", "Adult, …
$ individual_id <chr> "N1A1", "N1A2", "N2A1", "N3A1", "N3A2", "N4A1", "N4A…
$ clutch_completion <chr> "Yes", "Yes", "Yes", "Yes", "Yes", "No", "No", "Yes"…
$ date_egg <date> 2007-11-11, 2007-11-11, 2007-11-16, 2007-11-16, 200…
$ culmen_length_mm <dbl> 39.1, 39.5, 40.3, 36.7, 39.3, 38.9, 39.2, 41.1, 38.6…
$ culmen_depth_mm <dbl> 18.7, 17.4, 18.0, 19.3, 20.6, 17.8, 19.6, 17.6, 21.2…
$ flipper_length_mm <dbl> 181, 186, 195, 193, 190, 181, 195, 182, 191, 198, 18…
$ body_mass_g <dbl> 3750, 3800, 3250, 3450, 3650, 3625, 4675, 3200, 3800…
$ sex <chr> "male", "female", "female", "female", "male", "femal…
$ nitrogen <dbl> NA, 8.94956, 8.36821, 8.76651, 8.66496, 9.18718, 9.4…
$ carbon <dbl> NA, -24.69454, -25.33302, -25.32426, -25.29805, -25.…
$ comments <chr> "Not enough blood for isotopes.", NA, NA, NA, NA, "N…
$ year <dbl> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007…
$ mean_carbon <dbl> -25.81356, -25.81356, -25.81356, -25.81356, -25.8135…
$ mean_nitrogen <dbl> 8.859398, 8.859398, 8.859398, 8.859398, 8.859398, 8.…
$ sd_carbon <dbl> 0.5871106, 0.5871106, 0.5871106, 0.5871106, 0.587110…
$ sd_nitrogen <dbl> 0.4282233, 0.4282233, 0.4282233, 0.4282233, 0.428223…