[1] "/Users/dp415/Desktop/exeter_open_reproducibility_website"Making reproducible projects
An introduction to version control with R Studio and GitHub
& Matt Jones
20th June 2023
Workshop outline
- Workshop aims (reproducibility tools)
- Introduction to version control
- Overview of git and GitHub
- Using GitHub with R Studio
- Integrating with R Studio Projects and
here - Some use-cases from us
- Bonus session on data sharing! (w/ Matt Jones)
- Practical (bring your problem code!)
Three aims:

+

+
{here}
- Know basics of using git/GitHub for version control
- Learn how to create R Studio Projects
- Understand how the
herepackage works
Preparation
Checked git is installed
Signed up for a GitHub account
Authenticated GitHub on your machine



Let us know if you are stuck on any of these
(Extra guidance is on the Exeter data Analytics Hub)


Why use version control?
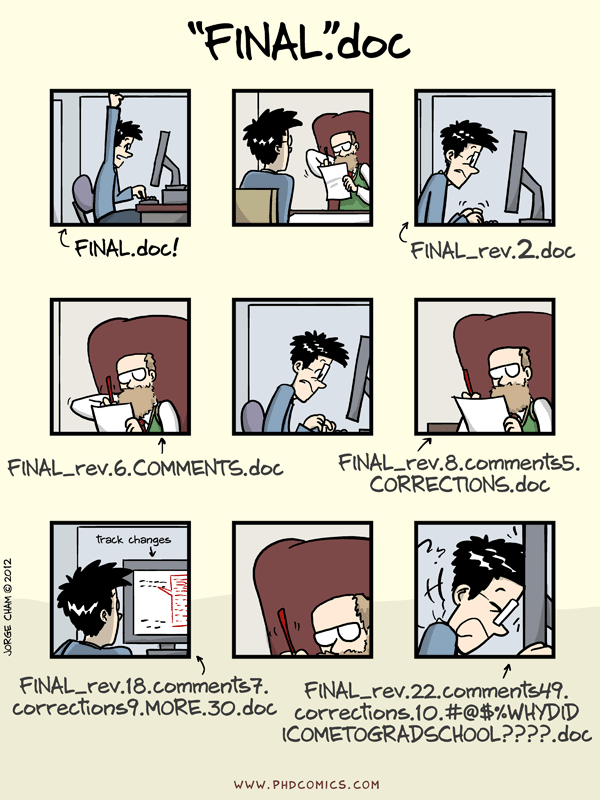
comic: “Piled Higher and Deeper” by Jorge Cham (www.phdcomics.com)
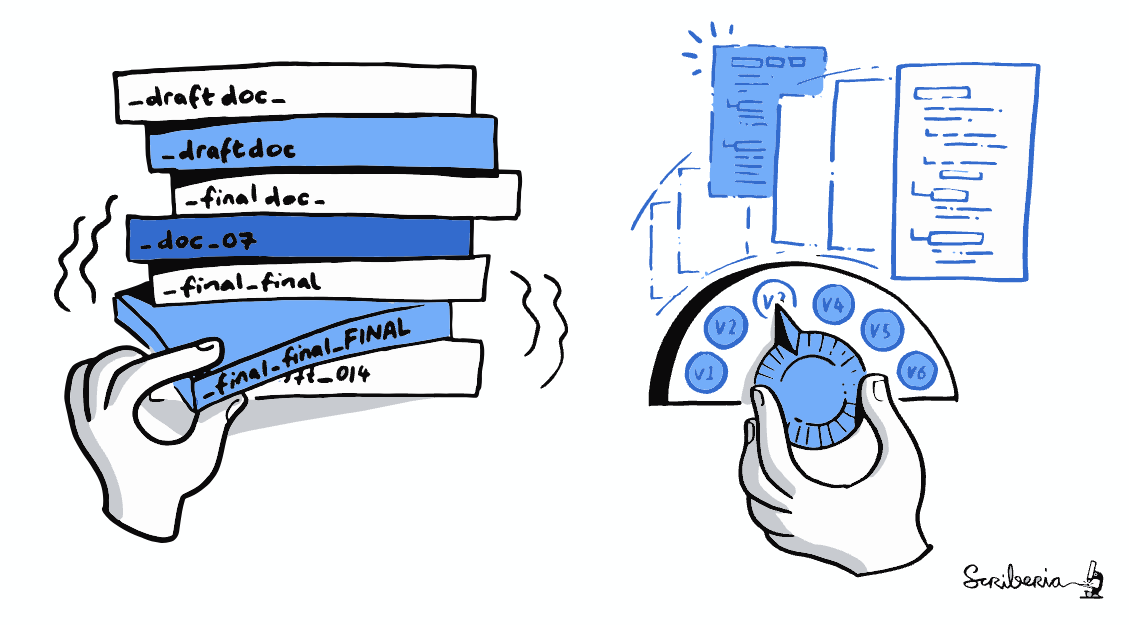
This illustration is created by Scriberia with The Turing Way community.
Used under a CC-BY 4.0 licence. DOI: 10.5281/zenodo.3332807
Version control allows us to:
- Avoid confusing file names
- Keep track of changes made over time
- Tinker with code without worrying about breaking it
- Easily revert when code does break
- Integrate with other software for online back-ups



- Installed locally
- Free version control system (often pre-installed)
- Manages the evolution of files in a sensible, highly structured way
- Structured around repositories (aka a ‘repo’) as units of organisation

- Cloud-based
- Hosting service for git-based projects (others: BitBucket, GitLab)
- Similar to DropBox/Google Docs but better
- Allows others to see, synchronise with and contribute to your work
The git/GitHub workflow
- Specific flow of actions that are usually followed:
Pull
Download everything from GitHub for the repo*
Stage
Add modified files to the commit queue

Commit
Confirm your changes locally (with message)

Push
Upload committed changes to GitHub

*Optional — but good practice to do when starting for the day
Interacting with git/GitHub


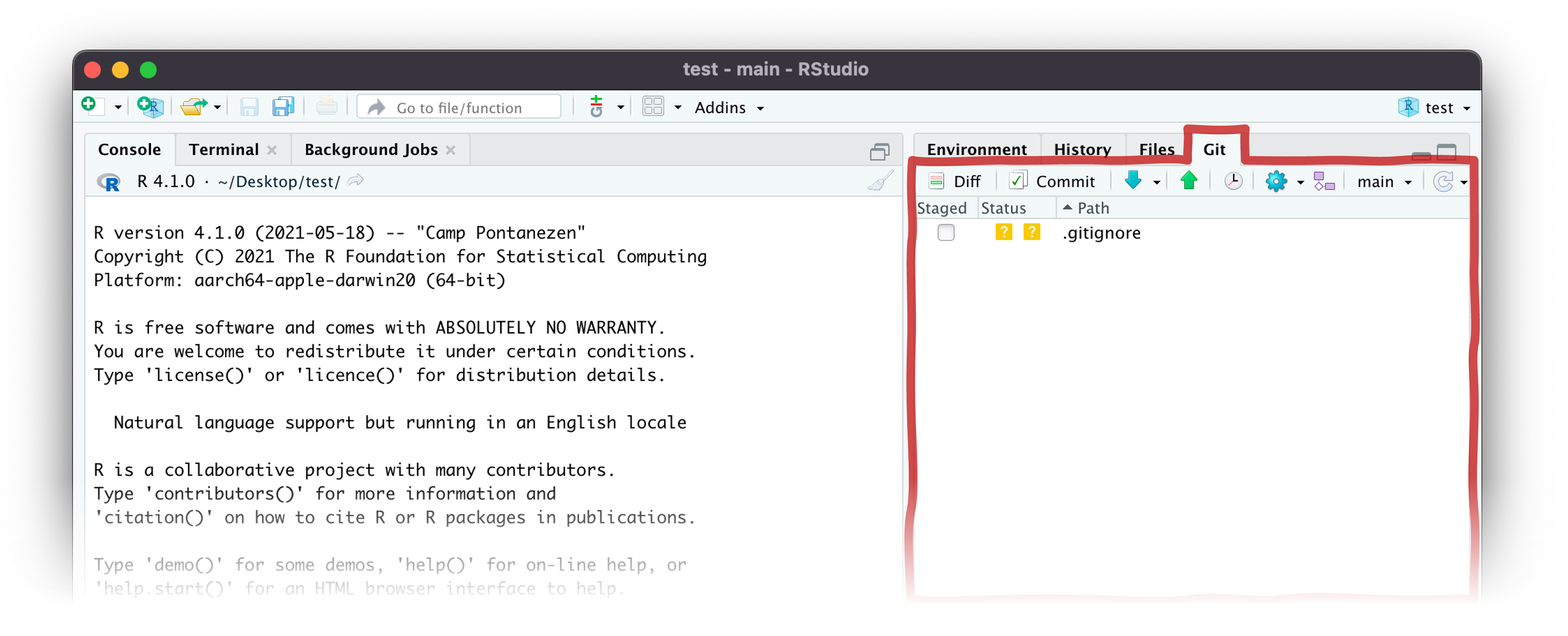
The R Studio git pane will only appear when you activate a project in version control
The git/GitHub workflow (RStudio)
- Same flow:
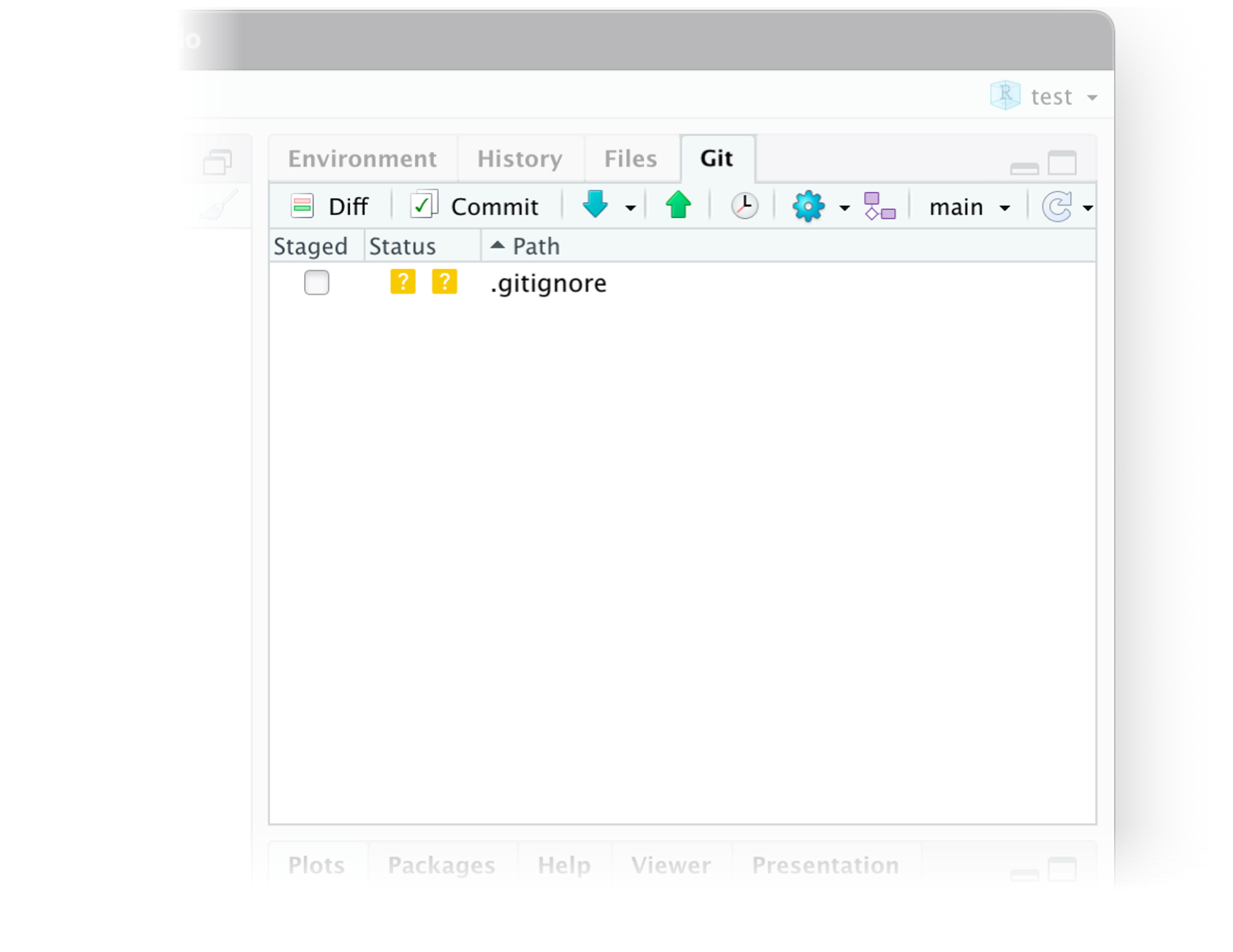
Pull

Stage


Commit


Push



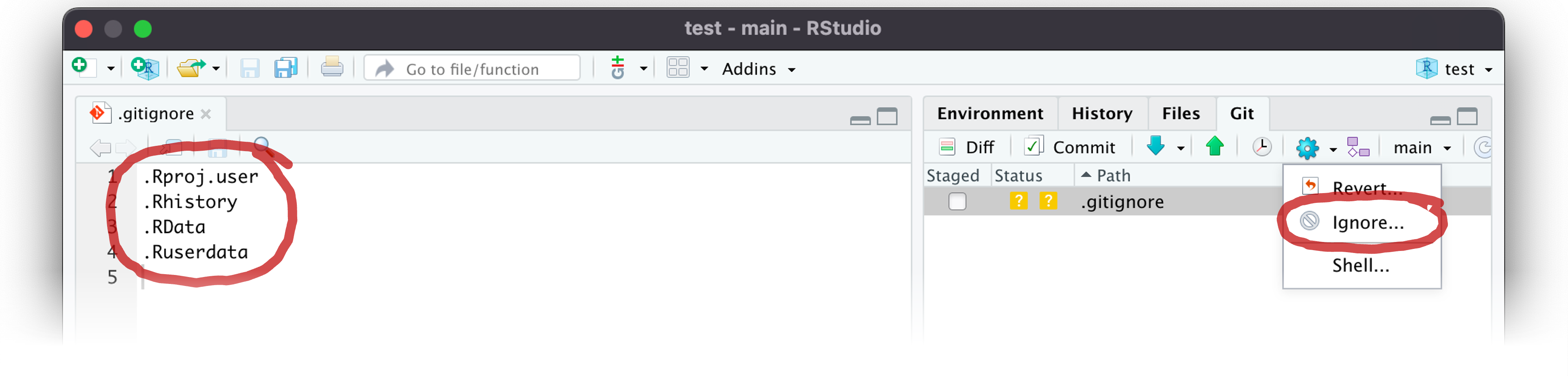
.gitignore

- Text file that lists large/specific files you don’t want to sync
- Exclude all files of one type with
*wildcard (e.g.,*.png)
- Edit the .gitignore file (left) or select files to exclude (right)

R Studio Projects
Why R Studio Projects are great:

- Each Project file opens a new session and environment
- File paths start relative to the .Rproj file (much shorter)
- Improves code reproducibility — even better if you use
here
- Self-contained project folder makes a perfect GitHub repo:

Guide to using R Studio projects can be found on the Exeter Data Analytics intros page
How to create an R Studio project
Guide to using R Studio projects can be found on the Exeter Data Analytics intros page
Create project by cloning a GitHub repo
Note: You can edit/save files from a cloned repo, but won’t be able to push unless you are the repo owner or a collaborator (details here)
{here}

Illustration by Allison Horst
{here} makes filepaths that:
- Work magically on both macOS & Windows (
/or\) - Start at the root of the repository (using .Rproj file)
here::here() will show the root of your project directory:
This path is included at the start of every filepath here creates
Recommended reading: Project-oriented workflows by Jenny Bryant
Using here:
- Filepaths are created in a similar way to
paste(): - We list quoted names of folders, comma separated
We can create a test filepath to a folder within the repository:
Create paths directly within a file/filename argument:
How I use GitHub:
- I usually start by making an empty repo on GitHub
- Clone the empty repo to my machine as an R Studio Project
- Create a few core folders (data, scripts, outputs)
- Add relevant files and commit-push everything
- Fire up a script and read in data with
here - Use
pacman::p_loadfor loading packages - …spend the next three hours faffing with ggplot
– Intermission –
Why you should share your data,
why you shouldn’t share it via GitHub,
and where you should share it instead.
Matt Lloyd Jones


Talk outline
- Why you should share your data
- Why you shouldn’t share your data via GitHub
- Where you should share your data instead
*Assuming you are permitted share your data
Talk outline
- Why you should share your data
- Why you shouldn’t share your data via GitHub
- Where you should share your data instead
*Assuming you are permitted share your data
You will soon have to share your data anyway.
- Concordat on Open Research Data
(signed by HEFCE, UKRI, Universities UK,
the Wellcome Trust and more1. - National Institutes of Health (NIH) has required
its fundees to eventually make their data
publicly available (as of January 2023)2. - US Government moving towards a position
of making sharing data mandatory
where possible)3.

Like sharing code, sharing data improves the quality of your science.
In the process of making your data
publication-ready, you will also
find yourself:
- Finding mistakes and correcting them
- Making sure the data inputted and
outputted from your code is consistent - Improving its documentation
(for future re-use - most likely by you!)

Your code won’t work without your data.
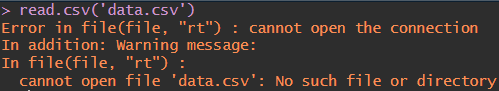
However, within reason, you shouldn’t store research data in your GitHub repository.
Talk outline
- Why you should share your data
- Why you shouldn’t share your data via GitHub
- Where you should share your data instead
You cannot assign a DOI to a GitHub repository.
- Like your publication, your data
should have a persistent identifier
like a Digital Object Identifier (DOI) - However, you can’t DOI your
GitHub repo, or versions of it! - For this reason, GitHub cannot be
considered a FAIR (Findable, Accessible,
Interoperable and Reusable) data repository

Memory limits
- Size of repository as a whole
cannot exceed 100 GB
(warnings >75 GB and >5GB)1,2 - Size of an individual push
(which may contain multiple
files) cannot exceed 2 GB1,2 - Size of each file in it cannot
exceed 100 MB (warnings > 50MB)1 - In order to prevent negatively
impacting GitHub’s infrastructure1

Inconveniences others
- Users who just want to play around
with your code are forced to download
all of your research data too
(potentially up to 100 GB!) - Smaller repositories are faster to clone
and easier to work with
git is not set up for handling data
- git version control system is based
around code, not data1 - git knows nothing about the structure of
common data formats we use
(e.g. the tabular structure of CSV files)2 - May result in merge conflicts
emerging where there are none2

GitHub is like your lab book, not to the freezer in which all your samples are kept.
Talk outline
- Why you should share your data
- Why you shouldn’t share your data via GitHub
- Where you should share your data instead
Rawness of data



Rawness of data
Things to consider when choosing somewhere to store your raw data
- Easy to download/upload
data from/to via code - Likely to stick around
- DOI-able

Example: Open Science framework
- Easy to download/upload
via theosfrpackage1 - Here to stay for the open
science revolution - Allows you to assign DOIs
to projects and/or datasets


Things to consider when choosing somewhere to store your processed data
- Higher memory limits
- Can store your final data in a
file structure (ideally alongside
the code that produced it) - DOI-able
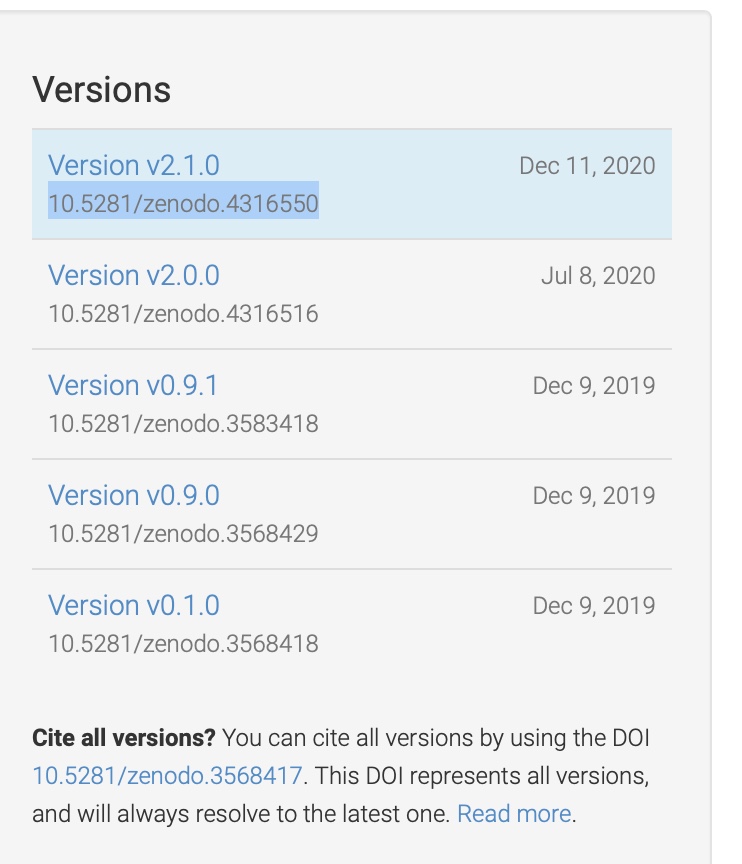
Example: Zenodo
- 50GB file size limit1
- You can just zip up your local version
of your Github repository (with both
code and data) at the end of running
all your code/analysis, and upload it - Allows you to assign a DOI to the
repository as a whole, as well as
to different versions of that repository
as it evolves through time
(and peer review) - Can also ‘reserve’ a DOI which is
really handy when writing a manuscript

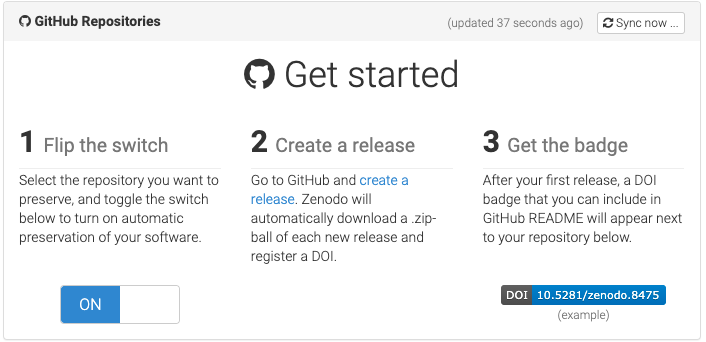
Zenodo example expanded
Would recommend NOT using the automated GitHub integration, because:
- Manual option allows you to reserve a DOI for use in submitted manuscripts (before making the dataset public)
- This only allows you to archive your code (since you’re not storing your data on GitHub anymore, right?)
- By zipping up and uploading the final, populated repository from your local machine, you can upload data and code together

Storing data outside of GitHub, but in a repositories friendly to GitHub keeps everyone happy!

![]()

Final considerations
- Following this schema, raw data will be archived on both OSF and Zenodo - but duplication is good in terms of data preservation
- OSF and Zenodo are generic repositories, but sometimes a more structured, subject-specific repository is required (e.g. NCBI or ENA for sequence data)
- You should consider whether your institution/funder/etc require you to also upload and store the data elsewhere
- You should consider whether you are allowed to share all of the data and/or whether you need to anonymise it (particularly raw data)
- You can prevent data from being uploaded (‘pushed’) to your GitHub repository alongside changes to code by storing it in data folders (e.g. ‘raw’ and ‘processed’ folders) and including these in your .gitignore file
Practical time!
How to spend the rest of the time:
Clone any GitHub repo (⭐️️)
Creating a new empty GitHub repo from scratch (️⭐⭐️)
Resources: happygitwithr - new GitHub project
Turn an existing project into a GitHub repo (⭐️⭐️⭐️)
Resources: (happygitwithr — GitHub first, GitHub last)
Feel free to ask us about your pesky code problems!
Resources
- ExeDataHub: Installing, R, RStudio, and Git
- ExeDataHub: Managing research projects with R Studio
- The exceptional Happy Git and GitHub for the useR book
- Others?
Acknowledgements
- UKRN for some original funding
- Various resources made by others

